Optimizing mRNA Vaccine & Therapeutic Production
Our Prima RNApols™ deliver higher yields and minimal double stranded RNA contamination compared to standard polymerases and can be tailored to your drug template and process. Prima RNApols™ have superior key performance indicators in the DNA–to–RNA in vitro transcription reaction that reduce manufacturing costs and improve mRNA quality.
Trusted by leading mRNA pharmaceutical partners, we have multiple collaborations using our standard Prima RNApols or developing a custom RNApol specific to their product, sequence, and/or process needs.
Contact us to learn more about our evaluation options or to discuss a custom polymerase program with our team.

Benefits of Prima RNApols™ Versus T7 Standard
Our novel RNA polymerases outperform T7 RNA polymerase (T7 RNApol). Prima RNApols™ are available for evaluation under an MTA, and we develop custom RNAP’s for specific needs.
Higher Yields of Target mRNA
Our process accommodates any DNA template sequence and length, enabling manufacturability of long mRNAs (>10 kb) for self-amplifying mRNA-based drugs, and higher yield per DNA template unit.
Improved mRNA Quality
Prima RNApols achieve up to a 100x reduction in dsRNA levels and minimizes other side-products, including short transcripts and truncated mRNAs.
Truly Enabling
Prima RNApols deliver higher performance and capping efficiency and increased nucleotide flexibility, resulting in higher IVT yield, lower manufacturing costs, and making previously non-manufacturable drugs viable.

Superior Quality
The difficulty of manufacturing high-quality mRNAs at large scale is the main challenge to the widespread use of mRNAs for other indications. The industry standard T7 RNA polymerase, used to synthesize mRNA from a DNA template in vitro, is inefficient and generates unwanted double-stranded RNA (dsRNA) byproducts that can trigger adverse immune responses. Our Prima RNApols overcome these obstacles by enabling production of mRNAs with higher yields, higher purity of target RNAs, and significantly lower dsRNA levels.
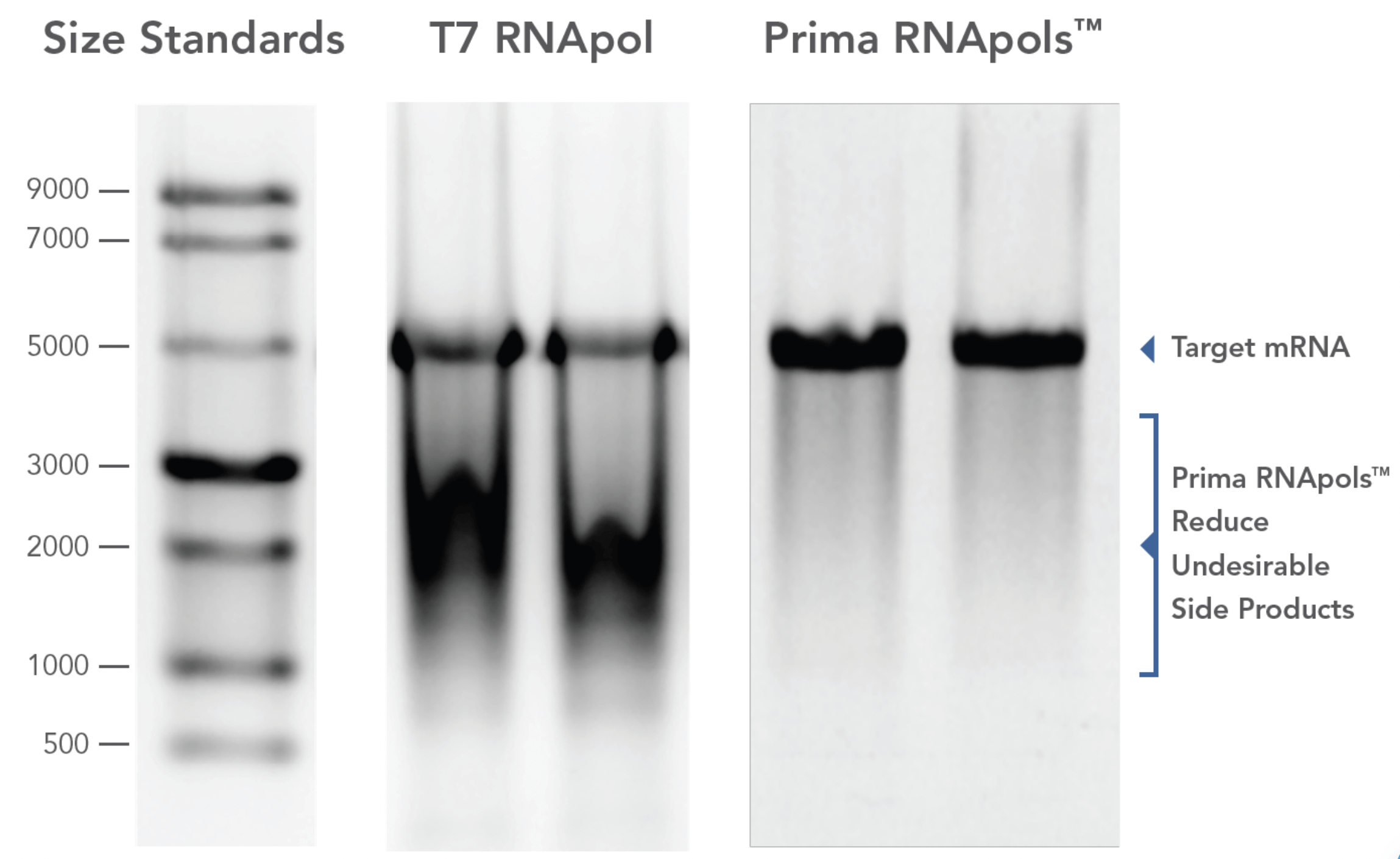
Gel image of in vitro transcription (IVT) reactions with a 5kb template.
Lane 1: single stranded RNA ladder; lanes 2 & 3: duplicate IVT reactions using T7 RNApol; lanes 4 & 5: duplicate IVT reactions using Prima RNApols. IVT reactions were treated with DNase I and a portion of the reaction was analyzed by agarose gel electrophoresis.
Performance Indicators
Prima RNApols™ KPIs (Relative to T7 RNApol)
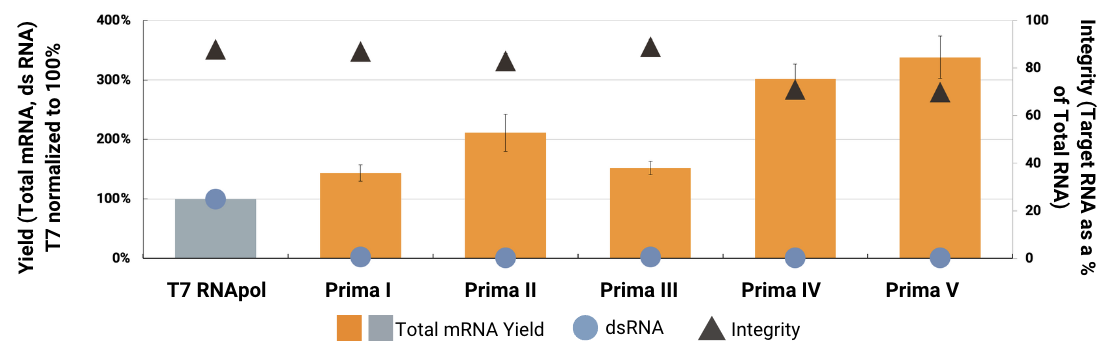
Total mRNA yield and integrity were measured on an Agilent Fragment Analyzer capillary electrophoresis system across all RNA sizes. The target yield was determined by quantifying the 5kb-sized peak. Integrity was calculated as the amount of the target RNA species as a percentage of total RNA. dsRNA levels were measured for equivalent amounts of total RNA synthesized in IVT reactions by immunoblotting with the J2 monoclonal antibody and quantitating spot intensities with ImageJ software.
mRNA Yield : Input Ratio
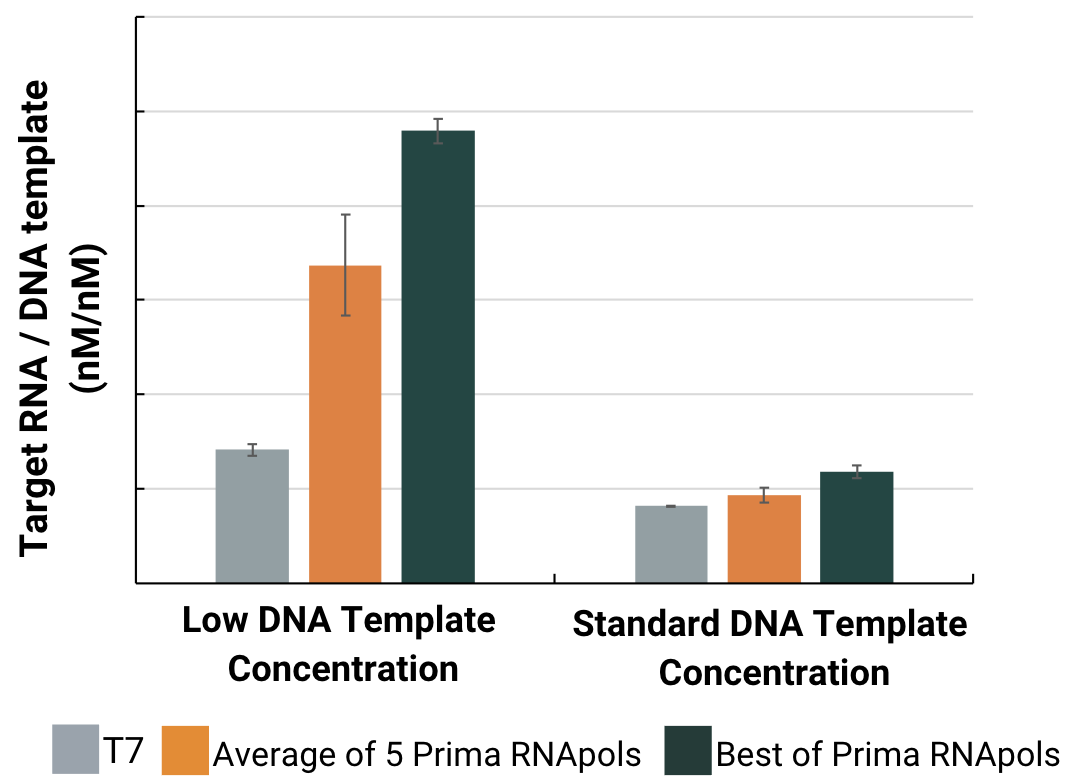
Total mRNA yield was measured on an Agilent Fragment Analyzer capillary electrophoresis system across all RNA sizes. The target yield was determined by quantitating the 5kb-sized peak.
Double-stranded RNA
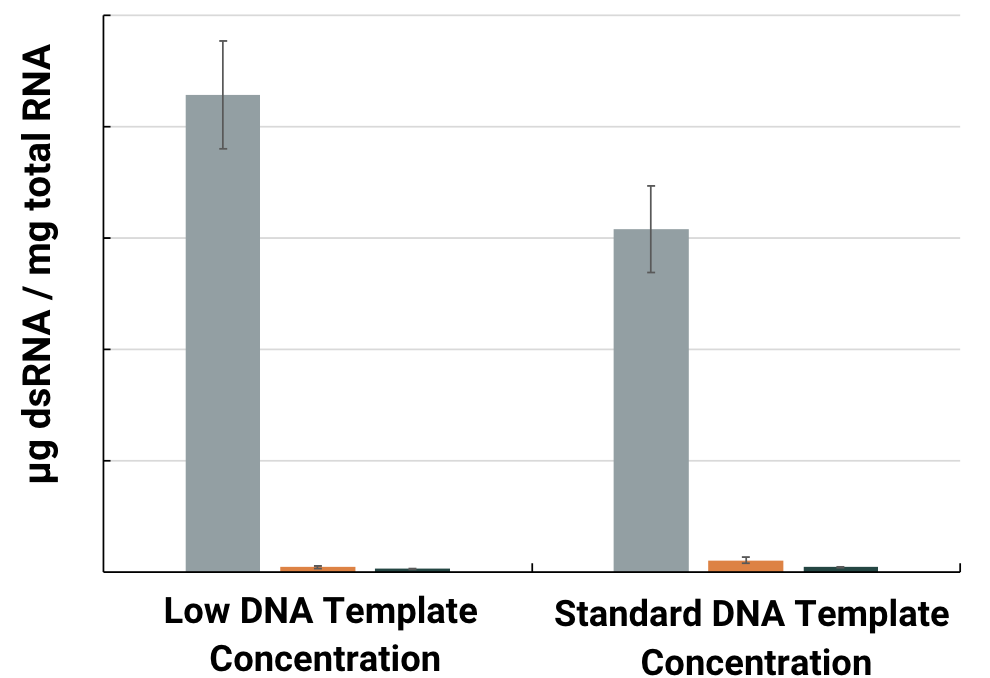
dsRNA levels were measured for equivalent amounts of total RNA synthesized in IVT reactions by ELISA using the SCICONS™ dsRNA ELISA Kit (based on the J2 monoclonal antibody).